Converting XML data to R dataframes with xmlconvert
The xmlconvert package is made to easily and comfortably convert XML data to R dataframes and the other way around. It provides a lot of options to control the conversion process and to export the results to CSV or Excel files, if desired.
How you install xmlconvert
Just execute install.packages("xmlconvert", dependencies = TRUE) in the R console to install the package including all packages it depends on. By default, the xlsx package for writing Excel files is not installed together with xmlconvert. xmlconvert will tell you when you need xlsx and will ask you to install it then.
How you work with xmlconvert
The functions xml_to_df() and df_to_xml() are the two workhorses of the package.
The functions assume that each data record (e.g. a customer) in the XML data is stored in one XML element/tag; the individual fields (e.g. name, address, e-mail), however, can be represented in different ways.
An XML data file could like this:
When working with the xml_to_df() and df_to_xml() functions, we would first of all use the records.tags argument to provide the tag name of the XML element that represents the records, in this example datarecord. Alternatively, the xml_to_df()’s records.xpath argument can be used to supply an XPath expression describing the location of the data records.
In the next step, we need to specify how to find the fields. In the example above, each field within a data record has its own XML element, and the tag name of this element is the field name. In this case, we would use fields = "tags" to make it clear to the xmlconvert functions that the fields are represented by XML elements. Not always will the tag names be the field names. The XML could also look like this:
Here, the XML elements representing the fields all have the tag field. The name of the field is not given by the tag name but is an attribute (called name in our case) of the field element. This attribute name can be supplied to the xmlconvert functions using the fields.names argument.
In this case, xml_to_df() could be called like this: xml_to_df("mydata.xml", records.tag = "datarecord", fields = "tags", fields.names = "name")
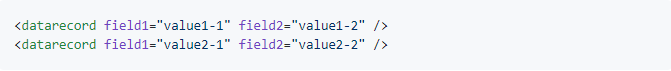
However, instead of being XML elements, the data fields could also be represented by individual attributes, as in the following example:
In this case, we use fields = "attributes" instead of fields = "tags" to adjust to this XML data structure. Here, a call of xml_to_df() could look like this: xml_to_df("mydata.xml", records.tag = "datarecord", fields="attributes")
The xmlconvert functions offer a lot of options to specify which the data fields are to be used, how to deal with missing values, how to treat with different data types and how to export the results. Please consult the online help by executing ?xml_to_df in the R console for more information on these options.
Contact the author
Joachim Zuckarelli
Twitter: @jsugarelli


Works like a charm! Any future plans on adding an argument to xml_to_df() concerning the maximum number of records to read?
ReplyDelete